ps <- readRDS("data/moving-pictures-ps.rds")Data Structure and Prevalence
Data structure and prevalence
Microbiome count data is structurally sparse and uneven.
This chapter focuses on:
- Matrix sparsity
- Library size variation
- Taxa prevalence
Load data
Sparsity
otu <- methods::as(phyloseq::otu_table(ps), "matrix")
if (!phyloseq::taxa_are_rows(ps)) {
otu <- t(otu)
}
zero_prop <- mean(otu == 0)
zero_prop[1] 0.9111154Library size variation
lib_size <- phyloseq::sample_sums(ps)
summary(lib_size) Min. 1st Qu. Median Mean 3rd Qu. Max.
897 1838 4010 4524 7013 9820 df_lib <- data.frame(
sample_id = names(lib_size),
library_size = as.numeric(lib_size),
stringsAsFactors = FALSE
)
dir.create("outputs/tables", recursive = TRUE, showWarnings = FALSE)
readr::write_csv(df_lib, "outputs/tables/library-size.csv")Library size distribution (Python)
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv("outputs/tables/library-size.csv")
fig, ax = plt.subplots(figsize=(8, 5))
ax.hist(df["library_size"], bins=25, edgecolor="white", linewidth=0.8)
ax.set_title("Library size distribution")
ax.set_xlabel("Library size (total reads per sample)")
ax.set_ylabel("Number of samples")
ax.grid(True, axis="y", alpha=0.25)
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
fig.tight_layout()
plt.show()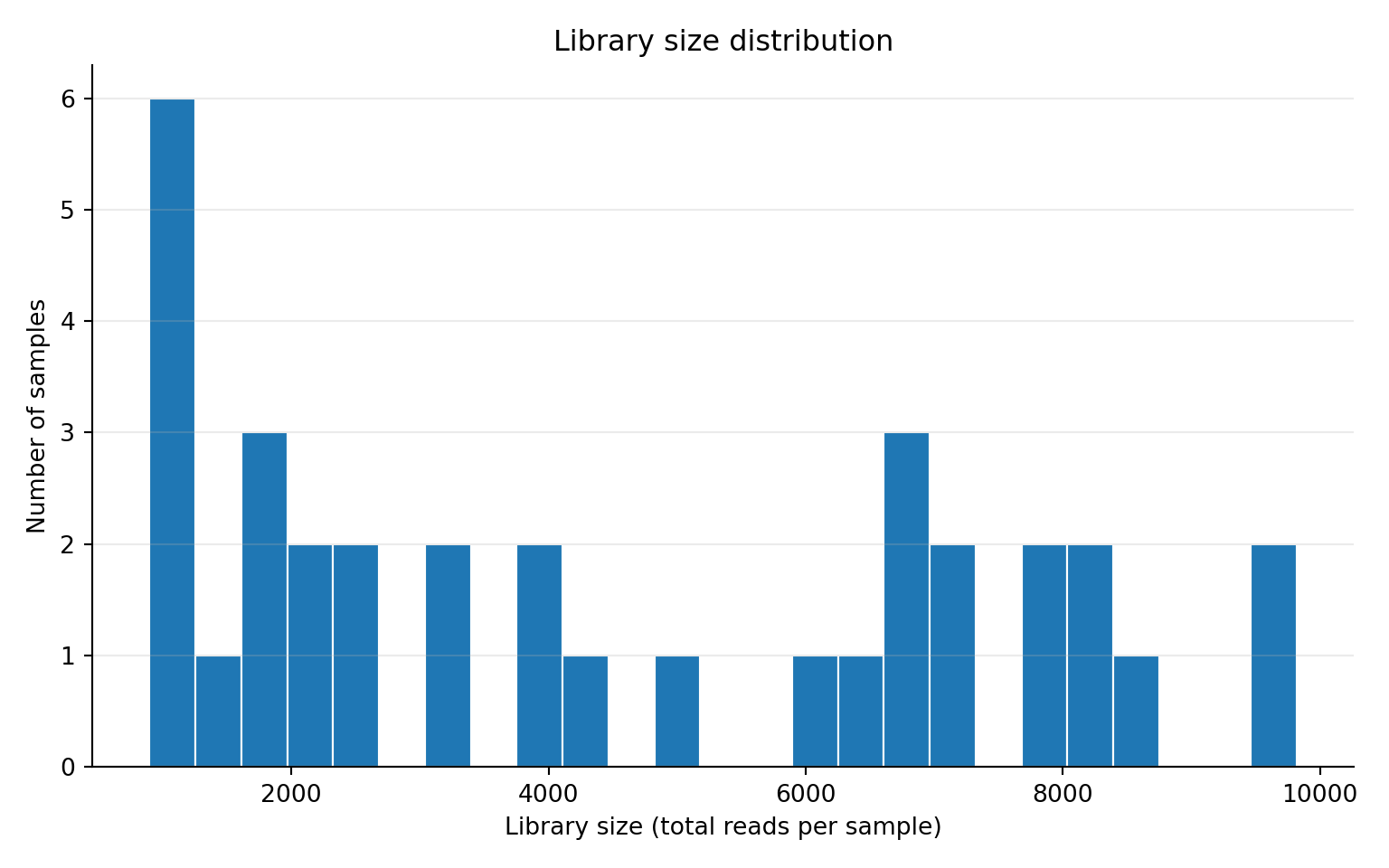
Detection and prevalence
detection <- 1
taxa_prev_n <- rowSums(otu >= detection)
taxa_prev_df <- data.frame(
taxon = rownames(otu),
prevalence_n = taxa_prev_n,
prevalence_frac = taxa_prev_n / ncol(otu),
stringsAsFactors = FALSE
)
summary(taxa_prev_df$prevalence_n) Min. 1st Qu. Median Mean 3rd Qu. Max.
1.000 1.000 2.000 3.022 4.000 26.000 dir.create("outputs/tables", recursive = TRUE, showWarnings = FALSE)
readr::write_csv(taxa_prev_df, "outputs/tables/taxa-prevalence.csv")Prevalence distribution (Python)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
df = pd.read_csv("outputs/tables/taxa-prevalence.csv")
fig, ax = plt.subplots(figsize=(8, 5))
ax.hist(df["prevalence_n"], bins=25, edgecolor="white", linewidth=0.8)
ax.set_title("Taxon prevalence distribution")
ax.set_xlabel("Number of samples detected")
ax.set_ylabel("Number of taxa")
ax.grid(True, axis="y", alpha=0.25)
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
fig.tight_layout()
plt.show()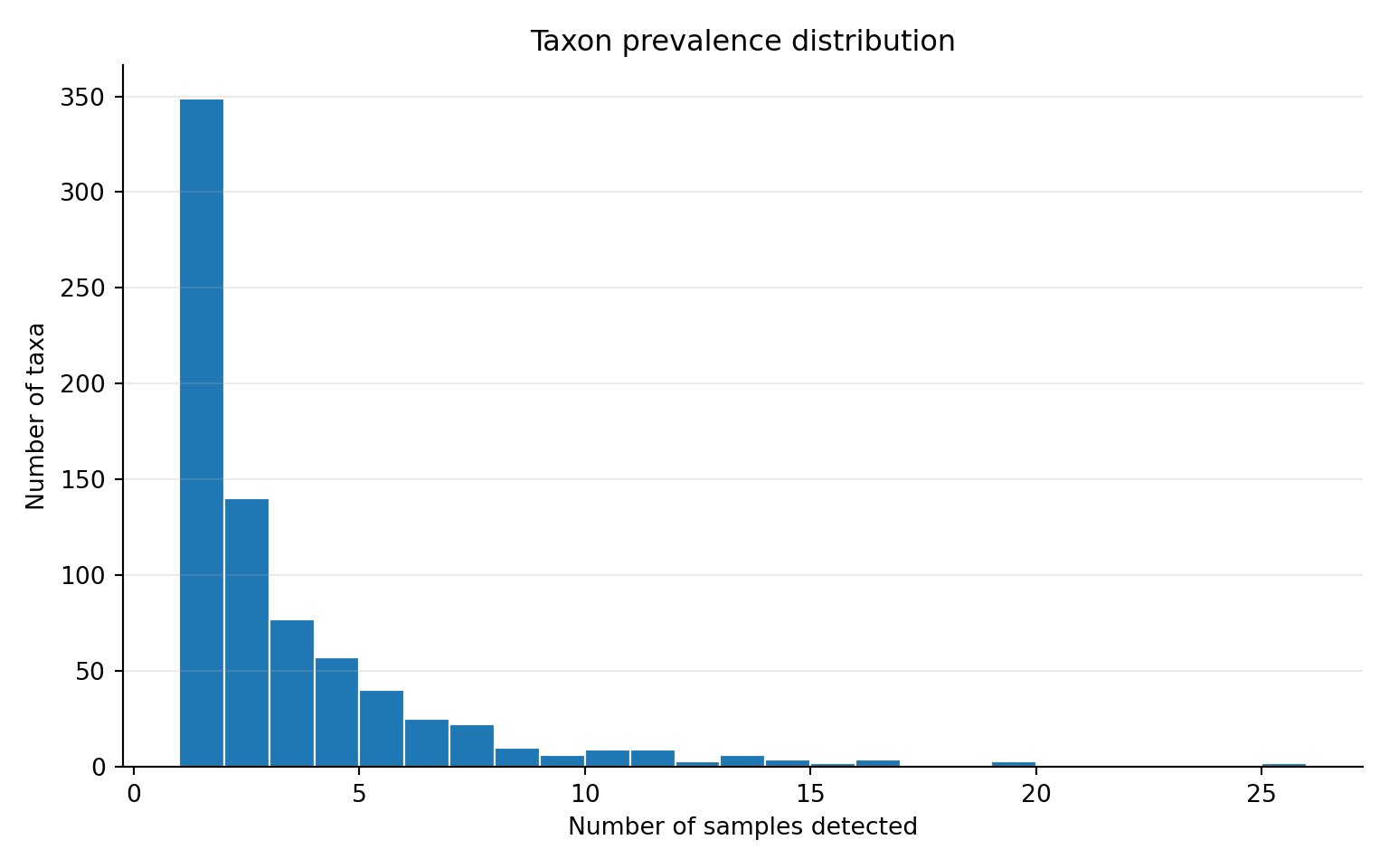
# Optional: ECDF view (often clearer for sparse data)
x = np.sort(df["prevalence_n"].to_numpy())
y = np.arange(1, len(x) + 1) / len(x)
fig, ax = plt.subplots(figsize=(8, 5))
ax.plot(x, y)
ax.set_title("ECDF of taxon prevalence")
ax.set_xlabel("Number of samples detected")
ax.set_ylabel("Fraction of taxa")
ax.grid(True, alpha=0.25)
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
fig.tight_layout()
plt.show()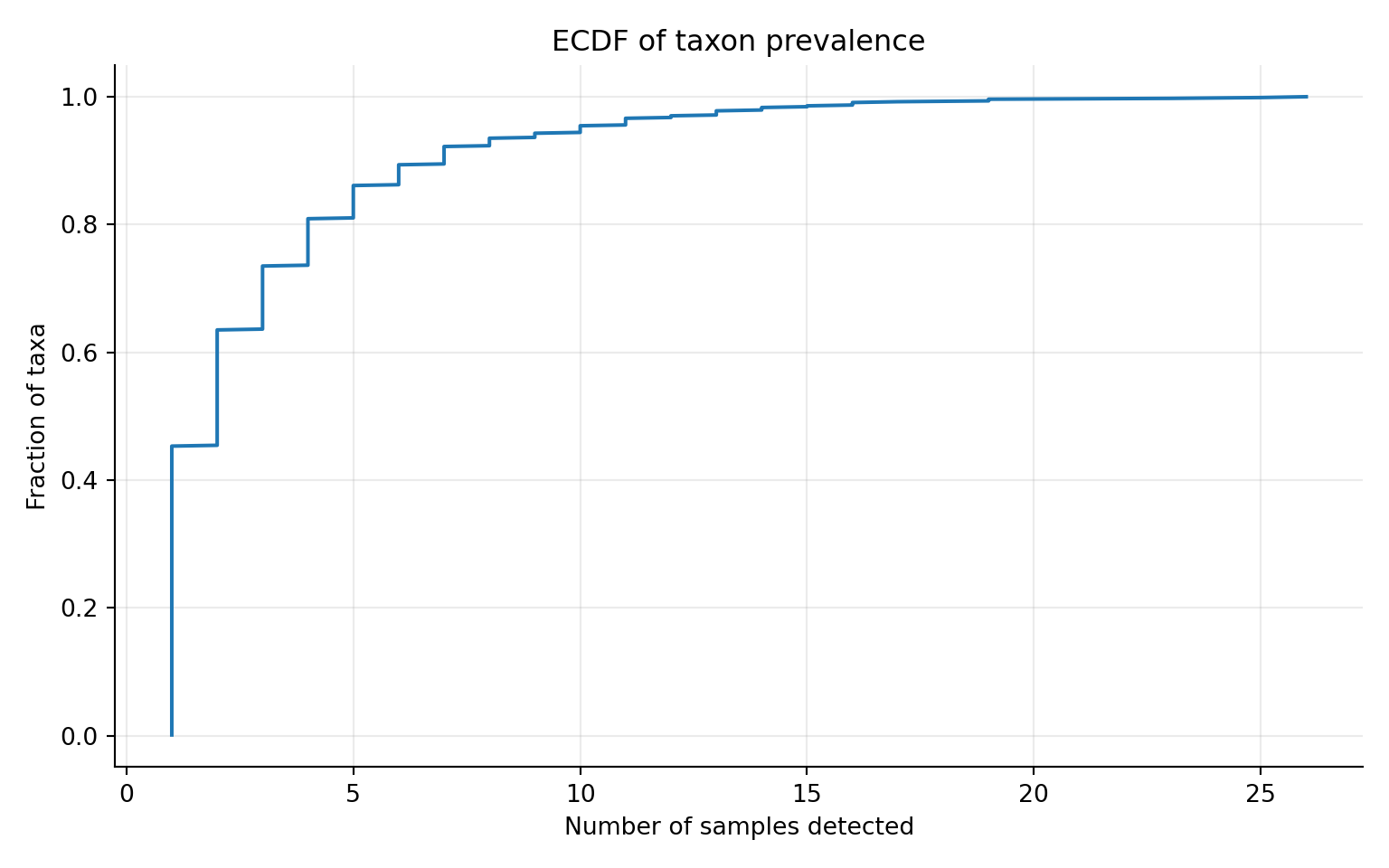
Simple prevalence filter
min_prev_frac <- 0.10
keep_taxa <- taxa_prev_df$taxon[taxa_prev_df$prevalence_frac >= min_prev_frac]
ps_prev <- phyloseq::prune_taxa(keep_taxa, ps)
c(original_taxa = phyloseq::ntaxa(ps), after_filter = phyloseq::ntaxa(ps_prev))original_taxa after_filter
770 204 Relative abundance preview
ps_rel <- phyloseq::transform_sample_counts(ps, function(x) x / sum(x))
range(phyloseq::sample_sums(ps_rel))[1] 1 1