library(phyloseq)
ps <- readRDS("data/moving-pictures-ps.rds")Heatmaps and Patterns
Heatmaps summarize many taxa across many samples at once.
They are useful for pattern discovery, but they are also easy to overread. A heatmap is a visualization of a transformed matrix, not a direct picture of biology.
This chapter focuses on:
- choosing a transformation appropriate for heatmaps
- selecting informative taxa (not just the rarest or noisiest)
- reading clustering and patterns responsibly
- avoiding common heatmap interpretation traps
Load data
Decide what to heatmap
Heatmaps work best when the matrix:
- has been transformed (raw counts are dominated by depth)
- has a limited number of taxa (readable signal)
- is aligned to metadata (so patterns can be interpreted)
We will create a genus-level heatmap using the top taxa by mean relative abundance.
Transform to relative abundance and aggregate to genus
ps_rel <- phyloseq::transform_sample_counts(ps, function(x) x / sum(x))
ps_genus <- phyloseq::tax_glom(ps_rel, taxrank = "Genus")
ps_genus <- phyloseq::subset_taxa(ps_genus, !is.na(Genus))
phyloseq::ntaxa(ps_genus)[1] 197Select top genera
genus_mean <- phyloseq::taxa_sums(ps_genus) / phyloseq::nsamples(ps_genus)
top_n <- 30
top_genera <- names(sort(genus_mean, decreasing = TRUE))[1:top_n]
ps_top <- phyloseq::prune_taxa(top_genera, ps_genus)
phyloseq::ntaxa(ps_top)[1] 30Build the heatmap matrix (R → Python)
We will export a clean matrix for Python plotting:
- rows: taxa (Genus)
- columns: samples
- values: centered log ratio style is premium-level; for free we use log1p relative abundance
We also export metadata to annotate columns.
# Extract matrix
mat <- methods::as(phyloseq::otu_table(ps_top), "matrix")
if (!phyloseq::taxa_are_rows(ps_top)) {
mat <- t(mat)
}
# Log transform for heatmap stability (free-track choice)
mat_log <- log1p(mat)
# Taxon labels
tax <- as.data.frame(phyloseq::tax_table(ps_top))
tax$taxon_id <- rownames(tax)
# Prefer Genus as label, fallback to taxon id if needed
label <- tax$Genus
label[is.na(label) | label == ""] <- tax$taxon_id[is.na(label) | label == ""]
# Apply labels
rownames(mat_log) <- make.unique(as.character(label))
# Export matrix
dir.create("outputs/tables", recursive = TRUE, showWarnings = FALSE)
mat_log_df <- tibble::as_tibble(mat_log, rownames = "taxon")
readr::write_csv(mat_log_df, "outputs/tables/heatmap-matrix.csv")
# Export metadata (for column annotation)
meta <- data.frame(phyloseq::sample_data(ps_top))
meta$sample_id <- rownames(meta)
# Robust body site detection
cols <- names(meta)
body_col <- intersect(c("body-site", "body.site", "body_site"), cols)
if (length(body_col) == 0) {
stop("Body site column not found in metadata. Available columns: ", paste(cols, collapse = ", "))
}
meta$body_site <- meta[[body_col[1]]]
meta_out <- meta[, c("sample_id", "body_site")]
readr::write_csv(meta_out, "outputs/tables/heatmap-metadata.csv")
# Quick sanity check
dim(mat_log)[1] 30 34Heatmap (Python, seaborn)
This is a modern heatmap with hierarchical clustering.
- Rows cluster by taxa similarity
- Columns cluster by sample similarity
- Body site is used as a column annotation
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
mat = pd.read_csv("outputs/tables/heatmap-matrix.csv", index_col=0)
meta = pd.read_csv("outputs/tables/heatmap-metadata.csv")
# Align metadata order to matrix columns
meta = meta.set_index("sample_id").reindex(mat.columns)
# Palette customization for annotations
palette_name = "Set1" # Try: "Set1", "tab10", "colorblind"
sites = meta["body_site"].astype(str).unique().tolist()
site_colors = dict(zip(sites, sns.color_palette(palette_name, n_colors=len(sites))))
col_colors = meta["body_site"].astype(str).map(site_colors)
sns.set_theme(style="white")
g = sns.clustermap(
mat,
method="average",
metric="euclidean",
col_colors=col_colors,
cmap="viridis",
figsize=(10, 9),
xticklabels=False,
yticklabels=True
)
# Legend for body site annotation
for site, c in site_colors.items():
g.ax_col_dendrogram.bar(0, 0, color=c, label=site, linewidth=0)
g.ax_col_dendrogram.legend(
title="Body site",
loc="center",
ncol=1,
bbox_to_anchor=(1.12, 0.5),
frameon=False
)
plt.show()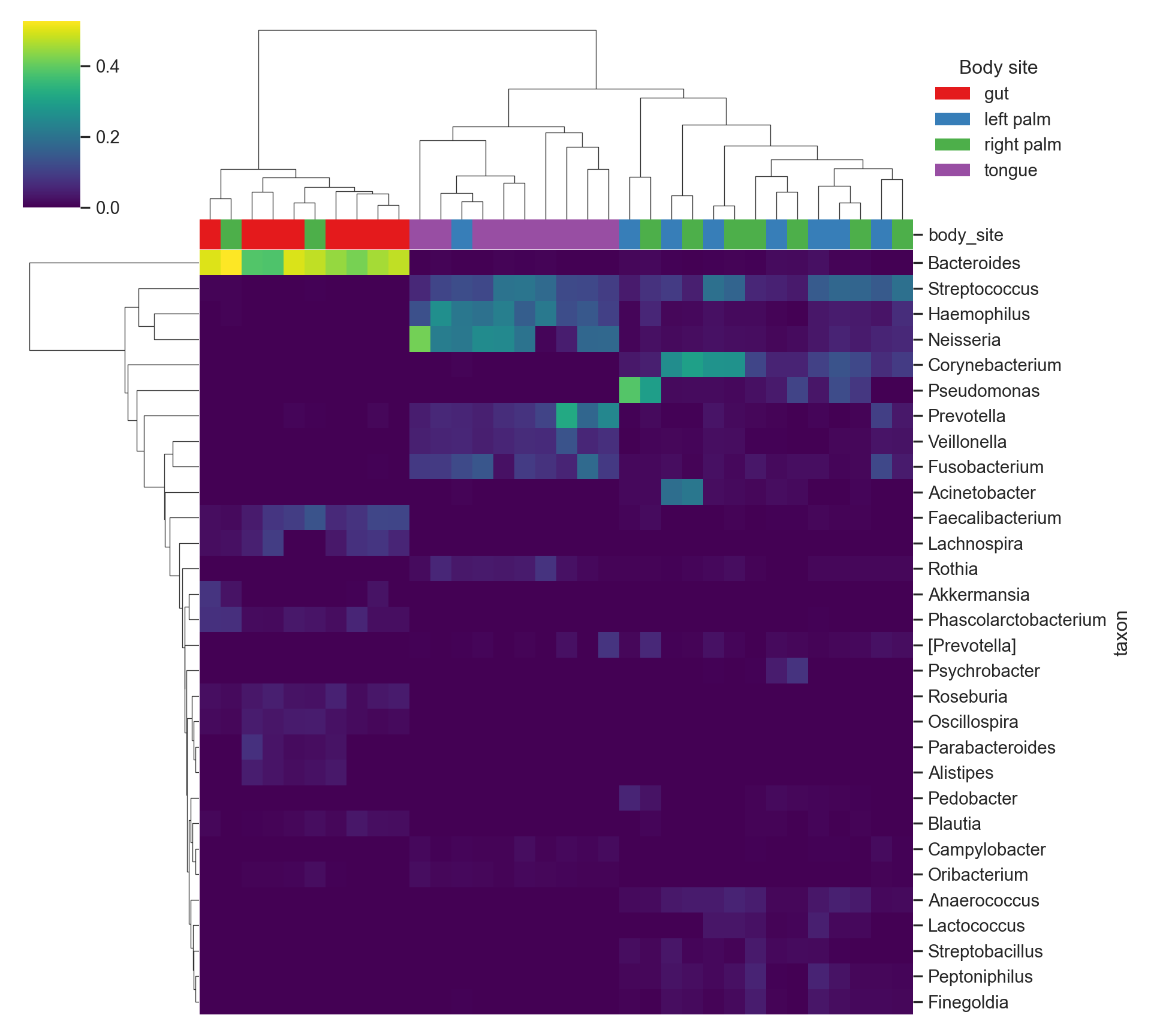
Reading heatmaps responsibly
What patterns can suggest
- Taxa that co-occur across samples
- Group structure (samples that cluster together)
- Dominance patterns (taxa that stand out repeatedly)
Common misinterpretations
- Clustering does not imply causation
- Bright color does not automatically imply biological importance
- Heatmaps are highly sensitive to:
- transformation
- filtering and top-N selection
- distance metric and linkage method