library(phyloseq)
ps <- readRDS("data/moving-pictures-ps.rds")Composition Visualization
Composition Visualization
Composition plots are often the first figures shown in microbiome studies.
Bar plots at the phylum or genus level are common. They appear intuitive. They are also easy to misinterpret.
This chapter focuses on:
- transforming counts to relative abundance
- aggregating taxa to a chosen rank
- visualizing composition across metadata groups
- understanding compositional limitations
Load data
Transform to relative abundance
Raw counts are not comparable across samples with different sequencing depth.
ps_rel <- phyloseq::transform_sample_counts(ps, function(x) x / sum(x))
range(phyloseq::sample_sums(ps_rel))[1] 1 1Aggregate to genus level
ps_genus <- phyloseq::tax_glom(ps_rel, taxrank = "Genus")
phyloseq::ntaxa(ps_genus)[1] 197Remove taxa without genus assignment:
ps_genus <- phyloseq::subset_taxa(ps_genus, !is.na(Genus))
phyloseq::ntaxa(ps_genus)[1] 197Identify top genera
To improve readability, select the globally most abundant genera.
genus_abundance <- phyloseq::taxa_sums(ps_genus)
top_genera <- names(sort(genus_abundance, decreasing = TRUE))[1:10]
ps_top <- phyloseq::prune_taxa(top_genera, ps_genus)
phyloseq::ntaxa(ps_top)[1] 10Prepare composition table for plotting
We will compute mean relative abundance by body site, then show two modern Python visualizations: a stacked bar chart and a heatmap.
# Long format table (sample-level)
df_comp <- phyloseq::psmelt(ps_top)
# Detect column names robustly (psmelt can vary by dataset)
cols <- names(df_comp)
sample_col <- if ("Sample" %in% cols) "Sample" else if ("SampleID" %in% cols) "SampleID" else if ("sample_id" %in% cols) "sample_id" else NA_character_
body_col <- if ("body-site" %in% cols) "body-site" else if ("body.site" %in% cols) "body.site" else if ("body_site" %in% cols) "body_site" else NA_character_
if (is.na(sample_col) || is.na(body_col)) {
stop(
"Expected columns not found in psmelt output.
",
"Available columns: ", paste(cols, collapse = ", "), "
",
"Missing: ",
paste(c(if (is.na(sample_col)) "Sample/SampleID" else NULL,
if (is.na(body_col)) "body-site/body.site/body_site" else NULL),
collapse = ", ")
)
}
# Keep only what we need and standardize names
df_comp <- df_comp[, c(sample_col, "Abundance", "Genus", body_col)]
names(df_comp) <- c("Sample", "Abundance", "Genus", "body_site")
# Mean relative abundance by (body_site, Genus)
df_mean <- aggregate(
Abundance ~ body_site + Genus,
data = df_comp,
FUN = mean
)
dir.create("outputs/tables", recursive = TRUE, showWarnings = FALSE)
readr::write_csv(df_mean, "outputs/tables/mean-composition-body-site-genus.csv")
head(df_mean) body_site Genus Abundance
1 gut Bacteroides 0.5623933731
2 left palm Bacteroides 0.0074999258
3 right palm Bacteroides 0.1497196754
4 tongue Bacteroides 0.0023039649
5 gut Corynebacterium 0.0001221555
6 left palm Corynebacterium 0.1273285793Composition by body site (Python stacked bar)
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv("outputs/tables/mean-composition-body-site-genus.csv")
# Pivot to body-site x genus
wide = df.pivot(index="body_site", columns="Genus", values="Abundance").fillna(0)
# Ensure consistent order (largest total first)
wide = wide.loc[wide.sum(axis=1).sort_values(ascending=False).index]
fig, ax = plt.subplots(figsize=(9, 5))
bottom = None
for col in wide.columns:
vals = wide[col].to_numpy()
if bottom is None:
ax.bar(wide.index, vals, label=col)
bottom = vals
else:
ax.bar(wide.index, vals, bottom=bottom, label=col)
bottom = bottom + vals
ax.set_title("Mean genus composition by body site")
ax.set_xlabel("Body site")
ax.set_ylabel("Mean relative abundance")
ax.grid(True, axis="y", alpha=0.25)
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
# Legend outside for readability
ax.legend(title="Genus", bbox_to_anchor=(1.02, 1), loc="upper left", frameon=False)
fig.tight_layout()
plt.show()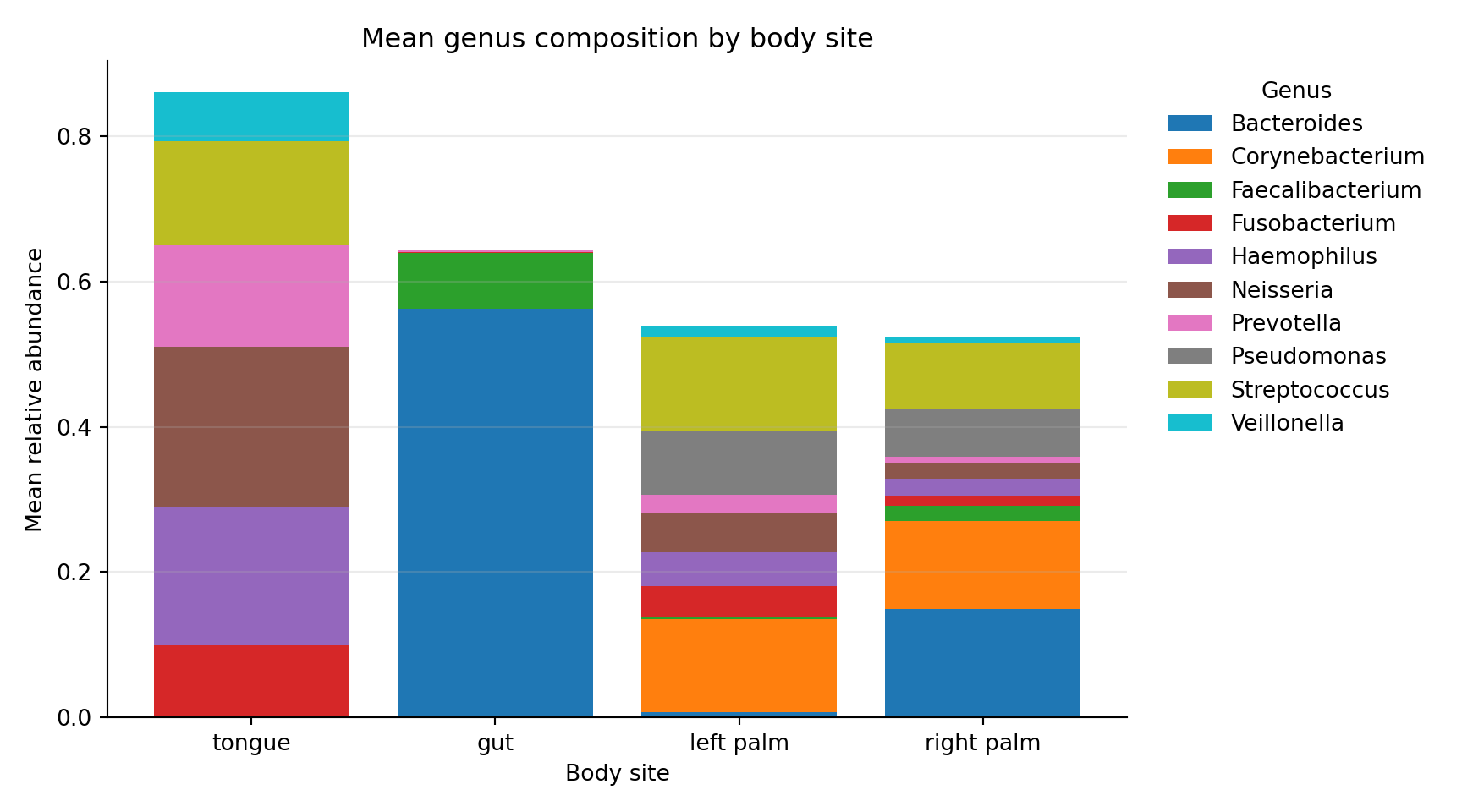
Composition heatmap (Python)
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv("outputs/tables/mean-composition-body-site-genus.csv")
wide = df.pivot(index="body_site", columns="Genus", values="Abundance").fillna(0)
# Order rows and columns by overall abundance
wide = wide.loc[wide.sum(axis=1).sort_values(ascending=False).index]
wide = wide[wide.sum(axis=0).sort_values(ascending=False).index]
fig, ax = plt.subplots(figsize=(9, 5))
im = ax.imshow(wide.values, aspect="auto")
ax.set_title("Heatmap of mean genus composition")
ax.set_xlabel("Genus")
ax.set_ylabel("Body site")
ax.set_yticks(range(len(wide.index)))
ax.set_yticklabels(list(wide.index))
ax.set_xticks(range(len(wide.columns)))
ax.set_xticklabels(list(wide.columns), rotation=45, ha="right")
cbar = fig.colorbar(im, ax=ax)
cbar.set_label("Mean relative abundance")
fig.tight_layout()
plt.show()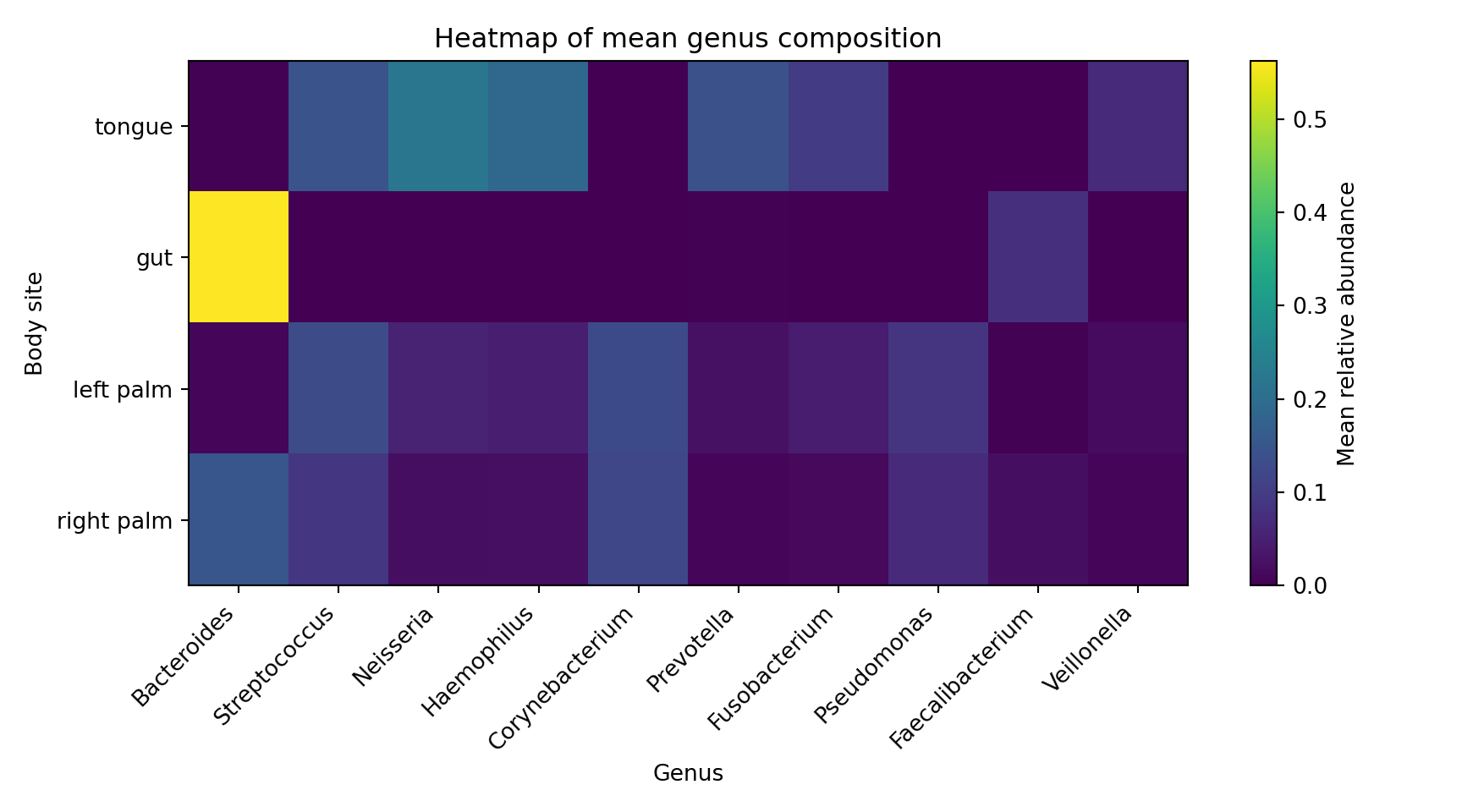
Limitations of composition plots
Composition plots:
- are constrained by constant-sum scaling
- cannot reveal absolute abundance changes
- are sensitive to filtering and aggregation level